IAM 950 HW1
Problem 1. Write numerical simulation for the heat eqn  on a periodic domain
on a periodic domain
![Math $[-L_x/2, L_x/2]$](/dokuwiki/lib/exe/fetch.php?media=wiki:latex:/img191e8ef8bbf8a610d84b39b39ce89df2.png) using Fourier spatial discretization and Crank-Nicolson temporal discretization.
Verify that your code works correctly by simulating the Gaussian-decay solution
using Fourier spatial discretization and Crank-Nicolson temporal discretization.
Verify that your code works correctly by simulating the Gaussian-decay solution

from t = 1 to 100 for parameters  , Lx=100, dt=1/16, Nx=128 (number of x gridpoints).
Initialize your numerical code with u(x,1) and then compare the results of the
numerical time integration with u(x,t) evaluated from the above formula. I suggest plotting
both quantities versus x at regular intervals in time during the simulation, using “pause” (matlab)
to stop the simulation temporarily.
, Lx=100, dt=1/16, Nx=128 (number of x gridpoints).
Initialize your numerical code with u(x,1) and then compare the results of the
numerical time integration with u(x,t) evaluated from the above formula. I suggest plotting
both quantities versus x at regular intervals in time during the simulation, using “pause” (matlab)
to stop the simulation temporarily.
Make a plot of the numerical solution and the Gaussian solution versus x at t=100 to turn in. Are the two entirely consistent? If not, why not?
Problem 2. Copy and revise your heat equation code so that it simulated the 1d Swift-Hohenberg equation,
 on a periodic domain
on a periodic domain ![Math $[0,L_x]$](/dokuwiki/lib/exe/fetch.php?media=wiki:latex:/img173b47829368ce70ed02ce3043a381e0.png) using Fourier spatial
discretization and Crank-Nicolson/Adams-Bashforth semi-implicit temporal discretization. Use the
initial condition
using Fourier spatial
discretization and Crank-Nicolson/Adams-Bashforth semi-implicit temporal discretization. Use the
initial condition  where
where  , parameter values
r = 0.2, Lx = 100, Nx = 256 (number of x gridpoints), dt = 1/16, and integrate from t=0 to 100.
, parameter values
r = 0.2, Lx = 100, Nx = 256 (number of x gridpoints), dt = 1/16, and integrate from t=0 to 100.
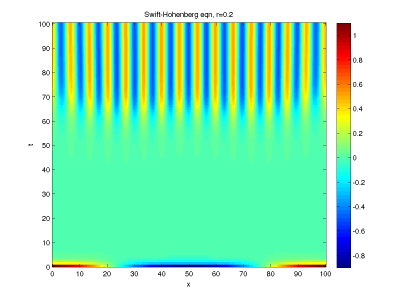
Make a colorplot of  in the
in the  plane (using every x gridpoint but plotting t at a larger
interval than dt, perhaps at intervals of 1/2 or 1. It should look something like this
plane (using every x gridpoint but plotting t at a larger
interval than dt, perhaps at intervals of 1/2 or 1. It should look something like this
When you are satisfied with the correctness of your simulation, make a plot of 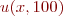 versus x for
the above parameters and initial conditions to turn in.
versus x for
the above parameters and initial conditions to turn in.
Now play around with the forcing parameter in the range 0 < r < 1 and the initial condition, by changing the wavelengths and magnitudes of the the cosines, and possibly the total integration time, if you don't see patterns develop by t=100. Answer the following questions
- Does the wavelength of the final pattern depend on r or the initial condition?
- Does the wavelength of the final pattern match you expectation from linear analysis?
- Can you relate the amplitude of the final pattern or the time scale of its growth from the initial condition to the Swift-Hohenberg equation?
Problem 3. Copy and revise your Swift-Hohenberg code so that it simulates the 1d Kuramoto-Sivashinsky equation,  , with parameters Lx = 100, Nx = 512, and dt = 1/16,
initial condition
, with parameters Lx = 100, Nx = 512, and dt = 1/16,
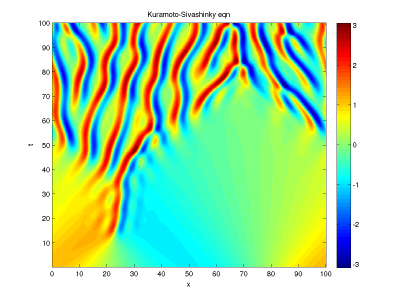
initial condition  (same as before), and integrate from t=0 to t=100. Make a colorplot of
(same as before), and integrate from t=0 to t=100. Make a colorplot of  as before. It should look like this
as before. It should look like this
Make a plot of 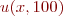 versus x to turn in. Now play around with the domain size Lx (adjusting
Nx so that Lx/Nx is approximately constant), try replacing the initial condition with random numbers
of order 0.01, and answer the following questions
versus x to turn in. Now play around with the domain size Lx (adjusting
Nx so that Lx/Nx is approximately constant), try replacing the initial condition with random numbers
of order 0.01, and answer the following questions
- Does the wavelength of the final pattern depend on r or the initial condition?
- Does the wavelength of the final pattern match you expectation from linear analysis?
- What qualitative changes in behavior do you observe as L increases from small (L=1) to large (L=100)?
Problem 4. For each of the above codes, fix the initial condition and parameters, and run the simulation with dt = 1/8, 1/4, …. What happens? Why?
Some help on Matlab's FFT: Matlab documentation is not very helpful about the ordering or normalization of Fourier coefficients in the FFT. However, a small numerical experiment clarifies. Let's construct a function with energy in known wavenumbers and observe where nonzero coefficients appear in the numerical FFT. I'll do 8 gridpoints for the periodic domain [0, 2pi].
>> x = 1/8*(0:7)*2*pi
x =
0 0.7854 1.5708 2.3562 3.1416 3.9270 4.7124 5.4978
>> k = 0;
>> u = cos(k*x) + 0.1*sin(k*x);
>> [real(fft(u)); imag(fft(u))]
ans =
8 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0
>> k = 1;
>> u = cos(k*x) + 0.1*sin(k*x);
>> [real(fft(u)); imag(fft(u))]
ans =
-0.0000 4.0000 0.0000 0 0.0000 0 0.0000 4.0000
0 -0.4000 0 -0.0000 0 0.0000 0 0.4000
>> k = 2;
>> u = cos(k*x) + 0.1*sin(k*x);
>> [real(fft(u)); imag(fft(u))]
ans =
-0.0000 -0.0000 4.0000 0.0000 0.0000 0.0000 4.0000 -0.0000
0 0 -0.4000 0 0 0 0.4000 0
>> k = 3;
>> u = cos(k*x) + 0.1*sin(k*x);
>> [real(fft(u)); imag(fft(u))]
ans =
-0.0000 0.0000 -0.0000 4.0000 0.0000 4.0000 -0.0000 0.0000
0 -0.0000 0.0000 -0.4000 0 0.4000 -0.0000 0.0000
>> k = 4;
>> u = cos(k*x) + 0.1*sin(k*x);
>> [real(fft(u)); imag(fft(u))]
ans =
0 0 0 0 8.0000 0 0 0
0 0.0000 0 0.0000 0 -0.0000 0 -0.0000
So the ordering of Fourier modes in the FFT for N=8 is k = [0 1 2 3 -4 -3 -2 -1] –not what I suggested it would be in class. The normalization is also different: the kth and -kth FFT coefficients for u(x) = cos(kx) are not 1/2, but N/2.
Also observe that the treatment of the highest-order Fourier mode k=-4 is a little weird, in that the 8 represents the sum of the k=4 and k=-4 cosine modes, with no representation of the k=4 and k=-4 sin modes. Without going into too much detail on this, Fourier spectral PDE codes typically just zero the highest-order mode at every time step. We can ignore this subtlety.
Based on the above, here's a short example of how to do Fourier differentiation in Matlab on a [0,L] periodic domain. This should be plenty to get you started for the time-stepping codes.
% A demonstration of Fourier differentiation in Matlab
% Parameters
L = 10;
N = 16;
x = L/N*(0:N-1);
% Set up u(x) and du/dx for some known L-periodic function
a = 2*pi/L
u = sin(a*x) + 0.2*cos(a*2*x);
dudx = a*cos(a*x) - 0.4*a*sin(a*2*x);
% Now compute dudx via FFT
k = [0:N/2-1 -N/2:-1];
alpha = 2*pi*k/L;
D = i*alpha;
dudx_fft = real(ifft(D.*fft(u)));
% Plot everybody
plot(x,u, 'b', x,dudx, 'g', x, dudx_fft, 'r.')
legend('u','du/dx','du/dx via FFT','location','southwest')
xlabel('x')